We are desperate to find a genetic cause, or at least a tractably small number of genetic causes of every trait we want to study. It's understandable that we want this kind of answer. Unfortunately, GWAS has not accounted for much of the heritability (estimated genetic fraction of causation) of most diseases and other similarly complex traits that have been studied, as we've often pointed out.
Rather than abandon the game, and especially since the dream of common variants having major effects on common disease (so they would be a usefully large market for Pharma to invest in targeting) is diminishing, we have had to become contortionists to try to find how or why genomics is still the way to approach such traits.
Our approach to this question is statistical and hence is based on repeated observation of enough observed instances of a variant for it to achieve statistical 'significance' in our data, whether or not that makes its effect of enough importance' for counter measures to be develop that are targeted against it. But this means that in any practical sense, we can't get large enough samples to detect the effects of very rare variants. We need other approaches.
One is to track variants in key gene regions among family members, looking for correlations between the presence of the variant and that of disease. How effective this will be depends on whether we know enough about the trait or about genes to find those variants that have such a track. If we find enough different variants in the same gene doing this in different families, that's strong evidence.
There are two ways, however, for rare variants to work. One, as just described, is for the variant to have a major effect all on its own. That could be detectable. But if combinations of many different rare variants are required, the variants coming from a number of genes and many different combinations having similar effects, this method may not work well. Unfortunately, there are theoretical reasons to think this will likely be the case.
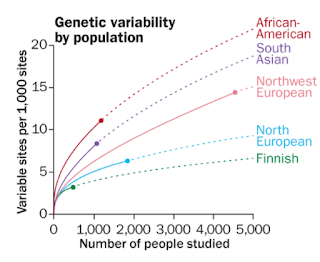
Recently there has been a story in Science News about the number of rare variants that we each carry around. The story summarizes various recent papers, citing the authors. The following graph shows the results of various studies of genome sequencing of different individuals:
This shows that each of us carries quite a few variants. The estimate is that there is about one variant per 1000 sites, if you compare both instances of the genome in a given person (or any two randomly chosen copies), which means about 3.1 million variants per pair compared. But the more people you look at the larger the number of sites that you'll see varying (even if the frequency of the rarer variant at the site is increasingly lower).
Obviously, in a huge population, almost any site will vary, and if lots of sites can potentially contribute to a disease, there will be lots of instances of 'causal' variants per gene, but these won't be detectable by group studies where one needs statistical association between the variant and the outcome (again, because statistical significance can't be achieved with very rare observations in this kind of study design).
The story was about disease hunting, which seems to be the international obsession (or, more accurately perhaps, rationale for funding the work). However, sites contributing to a trait's variation today will also potentially contribute to its evolution. Thus, in a subtle way too complex to go into here, hunting for the genes or variants that are responsible for the evolution of a trait is going to be very challenging, to say the least. It's hard enough to explain the selective (or chance) reasons for the trait's presence, much less what genes were responsible for its evolution.
The story also cites work by Andy Clark and Alon Keinan who have pointed out that the very rapid expansion of the human species in the 10,000 years since agriculture has generated a massive number of rare to very to very very rare variants. In a statistical sense, each gene lineage present at the beginning of that time has a million descendants today. This is not new or speculative theory but simply the consequence of the sequence-nature of genes: long strings of nucleotides mean many places where a nucleotide can change. Even if any given change is very rare, the genome and number of people born each generation are large. The variation is being found now that we can sequence at a high scale, as reports such as those mentioned here clearly show.
One sobering implication is that if we are concerned with the ways one can get a common disease or trait (behavior, morphology, or whatever, normal or not), then we face trying to work out this sea of nearly unique variation. It could be a hopeless task!
However, comparing close species with and without the trait in a sense aggregates the results of countless variants and genes and individuals over countless generations. In a subtle and statistically detectable way this could point to responsible genes , because the sample size that generated the result over time might leave enough evidence. Some methods to find such evidence are available (one test is called the Macdonald-Kreitman test, after its developers), though so far they are statistically rather weak for close species. Perhaps creative thinking will lead to new and better ideas.
Whether approaching disease in this way is the best thing to do is a separate question from how it can be done in practice if that is what, as currently, people are deciding they need to do.

5 comments:
Rather than missing heritability, its seems most of it is hidden below statistical significance. But even if you sequenced everyone on Earth, it would still be difficult to find a statistical association. If I were holding stock in a sequencing company, this situation would make me happy, but as a geneticist, I find this to be quite depressing.
Yes. We have said this in many of our posts, and in our series on probabilistic cause, posts on GWAS and so on, that you could probably find by searching on various keywords (like 'significance').
I agree with your conclusion. My response, and we've tried to say this in several posts, is that statistical significance is a holdover from 400 years ago, mainly developed for the physical sciences (and statistical testing for things like gambling), that may not apply to biology in the same way.
Maybe we need someone clever to think of better epistemological approaches that don't require assumptions of replicability, and where small causes are identified as being real.
And see our upcoming post tomorrow, on new papers about rare variants
Thanks for your response, Prof. Weiss. I've only recently started reading your blog and I really enjoy it!
Thank you, indeed! If you see other posts worthy of commenting on, please do.
Post a Comment